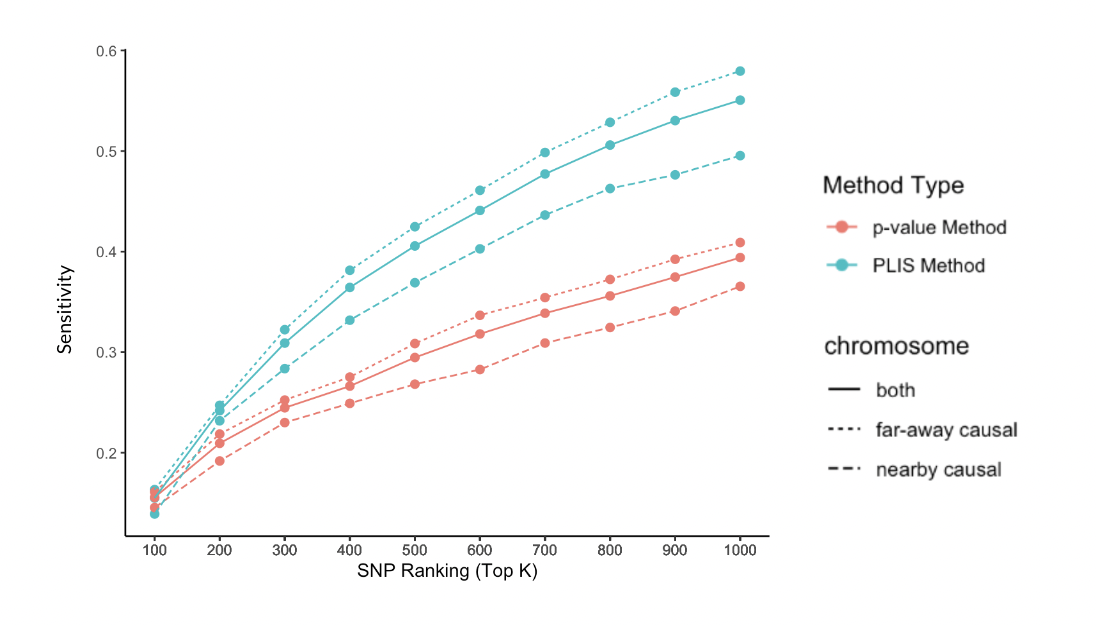
Together with my teammate Thu Dang, we learned from an article how to apply the Hidden Markov Model (HMM) to prune for linkage disequilibrium. Then, we replicated their results in R using HapMap data to showcase the efficacy of HMM. We additionally conducted a comparison of outcomes employing both traditional GWAS methodology and the Hidden Markov Model (HMM) approach for Type I Diabetes.
Linkage Disequilibrium Correction in Genome-Wide Association Study (GWAS)
Research
GWAS
R
This project uses Hidden Markov Model to correct for Linkage Disequilibrium in Genome-Wide Association Study.